Broadly describe the High Throughput Sequencing Workflow
8 - The High Throughput Sequencing Workflow
Sanger sequencing brought about a technological revolution, as it allowed to directly read DNA molecules with relative ease and affordability. The Human Genome Project motivated further progress, leading to automated DNA-sequencing machines capable of sequencing up to 384 samples in a single batch using capillary electrophoresis. Further advances enabled the development of high throughput sequencing (HTS), also known as next-generation sequencing (NGS) platforms.
At the moment, the high throughput sequencing technology most often used (by far) is Illumina. Similarly to the Sanger method, it is also based on the addition of nucleotides specifically modified to block DNA strand elongation, where each nucleotide is marked with a different color. Unlike the Sanger method, where a single DNA molecule is “read” at a time, modern illumina machines allow reading up to millions of DNA molecules simultaneously.
The following links are a good source of information regarding this sequencing technology:
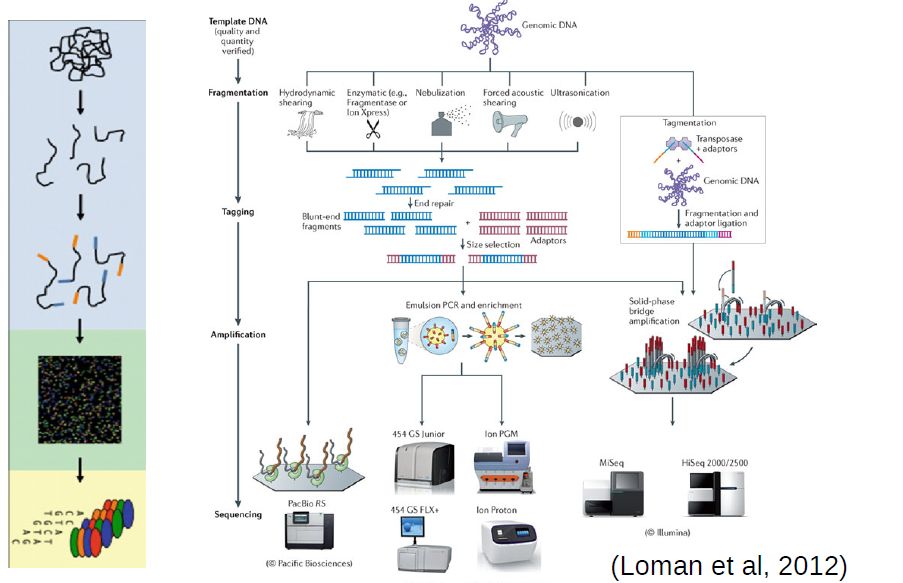
QUESTION: Can you broadly describe commmon steps in most high throughput sequencing workflows?
Click Here to see the answer
- Extraction and purification of the DNA template (even RNA must usually be converted to cDNA)
- Fragmentation of the DNA template (into a size range that can be accommodated by the machine)
- Attachment of sequencing tags (to enable reading by the machine)
- Amplification of signal (usually trough PCR, often already in the machine)
- Reading of signal and conversion into nucleotide bases
Back
Back to main page.