Quality control of label-free quantification MS data
By David Bouyssié
In this chapter, you will use a published DDA label-free dataset to learn some key aspects of quality control of such analyses. This will cover the quality of chromatographic alignment and of signal detection, as the well as the impact of shared peptides and data normalization on the final selection of significant hits.
Proteomic workflows based on nanoLC–MS/MS data-dependent-acquisition are now use routinely in core facilities and laboratories though popular software such as MaxQuant or Proteome Discoverer. However, the computational processing of label-free quantification data is challenging because of potential inconsistencies which can occur at several levels of the sample preparation and MS analysis. And more importantly mass spectrometers do not acquire MS/MS spectra in a reproducible manner in long series of samples, making mandatory to combine the identifications observed between the compared runs to obtain of full overview of the quantified peptides and proteins. Controlling the errors which may occur during the MS signal detection and the “match betwen runs” procedure is thus very important, since it can help to increase the reliability of quantitative studies based on this strategy.
In Ramus C. et al. J. Prot. 2016 , the authors set up a specific proteomic sample composed of an equimolar mixture of 48 human proteins (Sigma UPS1) spiked at different concentrations into a background of yeast cell lysate to benchmark several label-free quantitative workflows. Here we we will use a subset of the initial dataset to look at different quality control aspects.
1. General information
1.1 Experimental design
We will use two samples of 2 μg of a yeast (Saccharomyces cerevisiae) protein lysate spiked with two different concentrations of Sigma UPS1 (standard mixture of 48 equimolar human proteins). Each of these two samples was analyzed in triplicate on an LTQ Orbitrap Velos (160 minutes run duration). If you want to know more details about the sample preparation and the acquisition method please read the manuscript Experimental procedures.
:thought_balloon: Why did the authors combine two different Proteomes? What is the purpose of the Yeast background? What is the purpose of the set of 48 human proteins?
1.2 MS/MS search
Since the aim of this study was to compare software solutions the raw data were analysed by different pipelines. In this tutorial we will use the results obtained from the Mascot search engine. Here is a short description of the used parameters: Peaklists were submitted to Mascot database searches (version 2.4.2). ESI-TRAP was chosen as the instrument, trypsin/P as the enzyme and 2 missed cleavages were allowed. Precursor and fragment mass error tolerances were set at 5 ppm and 0.8 Da, respectively. Peptide variable modifications allowed during the search were: acetyl (Protein N-ter), oxidation (M), whereas carbamidomethyl (C) was set as fixed modification.
For more information regarding the Mascot parameters please check the corresponding documentation page.
The full list of raw files and result files are available from this PRIDE FTP site: ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2015/12/PXD001819
Two groups of UPS1-Yeast proteins were selected for this tutorial. Raw files and result files were renamed to work in blind conditions. Here is the list of files you will use:
| Group of samples | Raw file | Mascot file |
|---|---|---|
| G1 | OEMMA121101_36b | F083064.dat |
| G1 | OEMMA121101_38b | F083066.dat |
| G1 | OEMMA121101_40b | F083067.dat |
| G2 | OEMMA121101_61b | F083068.dat |
| G2 | OEMMA121101_63b | F083069.dat |
| G2 | OEMMA121101_65b | F083070.dat |
2. Visual inspection of LFQ results
Proline is a new software (manuscript under revision) for label-free workflow execution and raw data visualization. We will use the web interface of this tool to verify the quality of peak detection and LC-MS maps alignments.
For an introduction to the Proline algorithms implemnted for LC-MS peak detection and alignment please read the Proline algorithms documentation.
Additionally, you can also open the Web interface documentation before going through the rest of this tutorial.
2.1 Connection to Proline Web
Open a web browser (Chrome/Firefox) and go to: http://134.158.247.163:443
To log in, use as login/password userX/userX, where X is a number from 1 to 9.
Click on the button Apps then Dataset Expolorer.
2.2 Browse the results
In the “Yeast-UPS1” project expand the node called “Identification Trees” then double click on the “Yeast-UPS1” dataset. On the right hand side
| Raw file | Result file | #Val. protein sets | #Val. peptides | #Queries |
|---|---|---|---|---|
| OEMMA121101_36b | F083064.dat | 709 | 3535 | 37691 |
| OEMMA121101_38b | F083066.dat | 687 | 3500 | 37831 |
| OEMMA121101_40b | F083067.dat | 694 | 3442 | 37816 |
| OEMMA121101_61b | F083068.dat | 665 | 3708 | 41639 |
| OEMMA121101_63b | F083069.dat | 675 | 3850 | 41458 |
| OEMMA121101_65b | F083070.dat | 681 | 3904 | 41500 |
The number of queries corresponds to the number of MS/MS spectra which were submitted for the search.
Some quick definitions:
- each MS/MS spectrum may be identified by one ore multiple peptides. Each pair of “peptide <-> spectrum” is called a “Peptide Spectrum Match” (PSM), sometimes referred as “Peptide Match” in the Proline software.
- a peptide is a sequence of amnio acids plus a list of post-translational modifications localized on this sequence.
- protein sequences identified from the database by a same set of peptide sequences are grouped into entities called “Protein sets”.
:thought_balloon: Why the number of identified peptides and protein sets is different between the triplicates of the same experimental group? Do you see significant differences between the two groups (G1&G2)? Can you draw a conclusion from these results?
ANSWERS
2.3 Comparing human protein identifications
Double click on one of replicate of the group 1 then open the Proteins tab. On the left hand side of the first table expand the filters and in Text data add an Accession field, type HUMAN then click on Apply.
:thought_balloon: How many human proteins have been validated?
Click on the magnifier button before the “ALBU_HUMAN_UPS” accession. This should open a new page with detailled information about identified peptides for this protein.
Repeat the same procedures for one of the replicates of the group 2.
:thought_balloon: From these results, do you have an idea of the group having the highest concentration of UPS1?
ANSWERS
2.3 Quality control of quantitative results
In the “Yeast-UPS1” project expand the node called “Quantitations” then double click on the “Yeast-UPS1” dataset.
2.3.1 LC-MS maps alignments
Go to the tab called LC-MS maps, then you should see a plot similar to this one:
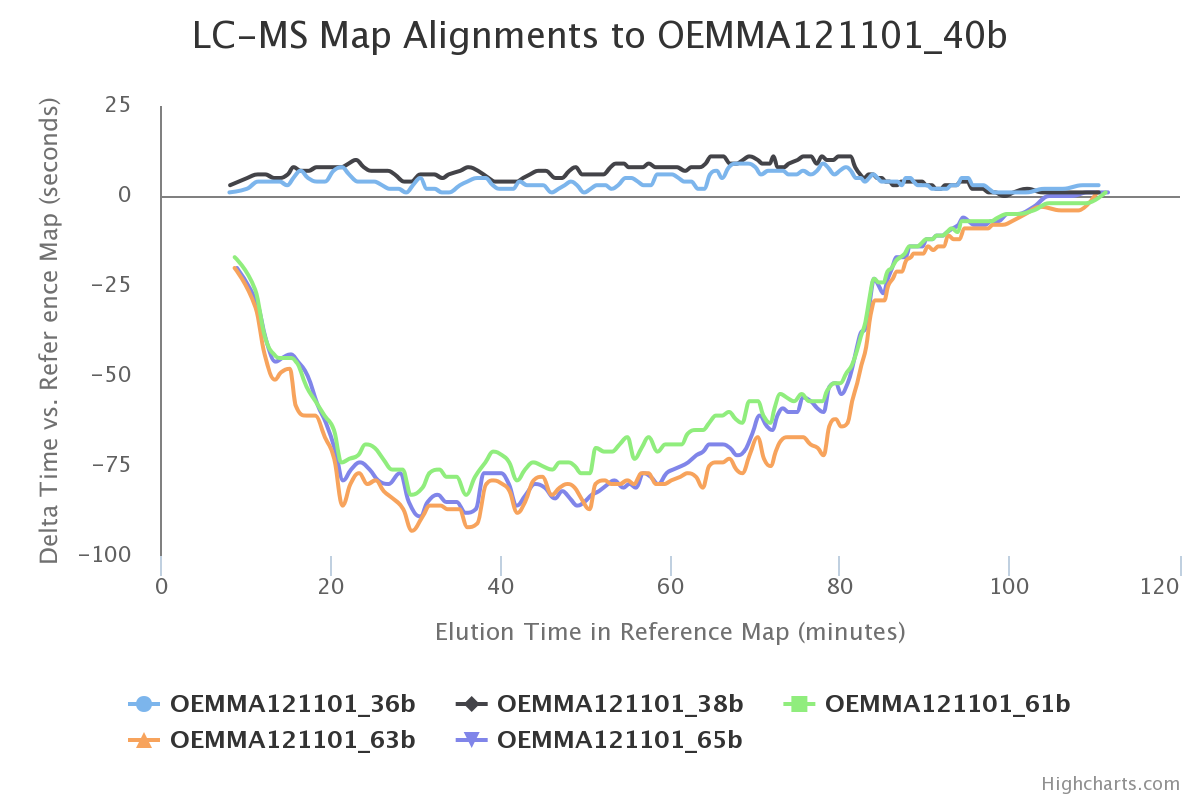
This plot correspond to the result of the LC-MS alignment which was performed by software. It help us to verify if there was any issue regarding the chromatographic reproducibility. The lowest retention time deviation, the better results we have.
:thought_balloon: Why does the plot contain only five curves? Do you think these results are consistent according to the experimental design? Can you use this plot to define the retention time tolerance for the “match between runs” procedure of the LFQ workflow (note: this procedure is called “cross-assignement” in the Proline documentation)?
ANSWERS
2.3.2 Volcano plots
Go to the tab called Quantitation Stats to visualize the volcano plot at protein level.
On the bottom left use the filters to highlight human proteins: click on a color in the palette then in the Accession field type HUMAN.
Set the T-Test p-value filter to 0.01, then sort the table in the upper left by the first column (Sel.).
:thought_balloon: How many proteins are selected using the T-Test filter? If we consider that the 48 human proteins were detected as significant results, how many yeast proteins are selected by this procedure? How can you explain thes results? Do you have any suggestion to improve them (look at the shape of the Volcano plot)? Try to fix it using the software.
ANSWERS
2.3.2 Verification of corresponding raw data
On the volcano plot the protein named RL8B_YEAST seems to be an outlier. Click on this point to open the detailled quantitative results.
:thought_balloon: Only 3 peptides are selected while 17 were quantified? Can you explain this result after having checked the parameters in “Summary” -> “Stats Params”?
ANSWERS
Double-click on the peptide named “AKNPLTHSTPK”. Compare these chromatograms with those obtained for the other selected peptides. You can also click on the “blue eye” button to perform manual XICs and check the signal detection.
:thought_balloon: For the 2+ charge state of “AKNPLTHSTPK” are you able to recover the XICs by performing a manual analysis? How could you explain the differences in intensity we observe for this peptide between the two compared groups? Would you keep this peptide to estimate the abundance of the protein?
ANSWERS
Go back to the proteins tab and search for the protein named “LSB3_YEAS7”. This protein is absent from one of the group
:thought_balloon: This protein seems to be absent from one of the groups. Perform a manual XIC to verify if it’s true. How could you explain these results?
ANSWERS
2.3.3 More advanced QC & statistics
In the “Exported files” node of the project tree, download the .xlsx file on your computer. We will now use some R scripts to generate and inspect quality control reports.
TODO: add links to results from Coralie’s scripts
Back
Back to main page.