Application of PSMC to MS-simulated data
This is a simple tutorial showing how to use PSMC to infer the population size history from full DNA sequences. We will apply the PSMC to simulated data.
We need:
- The ms software.
- The PSMC software.
- Two python scripts (one for converting files and the other one for plotting the results)
Downloading and compiling softwares
Download and compile ms
- Go to http://home.uchicago.edu/rhudson1/source/mksamples.html and download the file ms.tar.gz
- Uncompress the file
tar -zxvf ./ms.tar
- Compile ms
cd msdir
gcc -O3 -o ms ms.c streec.c rand1.c -lm
Have a look to the ms readme file for more details. The original paper can be viewed here
Download and compile psmc
- Clone the github repository
git clone https://github.com/lh3/psmc.git
- Compile
cd psmc
make
cd utils
make
The model of the PSMC was published by [Li and Durbin][1] in 2011. For the analysis we will generate a dataset similar to the first dataset used in the Supplementary Materials of the Li and Durbin paper.
Download the python scripts
git clone https://github.com/willyrv/ms-PSMC.git
Finaly, create a new folder and copy the binary files ./msdir/ms and ./psmc/psmc as long as the python scripts.
Simulating data and running the analysis
I will suppose you have all this file in a folder:
|- ms
|- psmc
|- ms2psmcfa.py
|- plot_results.py
You need to verify that the files have execution permissions. To give execution permissions to the files type:
chmod +x ./*
1-) Simulate the data with ms. Run:
./ms 2 100 -t 30000 -r 6000 30000000 -eN 0.01 0.1 -eN 0.06 1 -eN 0.2 0.5 -eN 1 1 -eN 2 2 -p 8 > sim1.ms
Normally this takes less than 30 seconds and should produce one output file (here called “sim1.ms”). Now we will use the ms2psmcfa.py python script to convert this ms output into the input file of PSMC.
2-) Run:
./ms2psmcfa.py ./sim1.ms > sim1.psmcfa
This creates the file “sim1.psmcfa” which is the input of PSMC. (This step is also fast, less than 30 seconds). Now we can run the PSMC analysis. For explanations about the parameters, see the README file of PSMC.
3-) Run:
./psmc -N25 -t15 -r5 -p 4+25*2+4+6 -o dem_history_sim1.psmc ./sim1.psmcfa
This is a bit long (about 3 hours). At the end, you should get a file like this
If you don’t want to wait, you can download the result file
wget http://wrodrigu.perso.math.cnrs.fr/GTPB/tutorial_psmc/result_files/dem_history_sim1.psmc
Once the psmc analysis is finished, you can plot the results.
4-) Run:
./plot_results.py
And you will finally get a figure with the inferred demographic history (dashed blue line) and the real history (continuous black line) corresponding to the ms command.
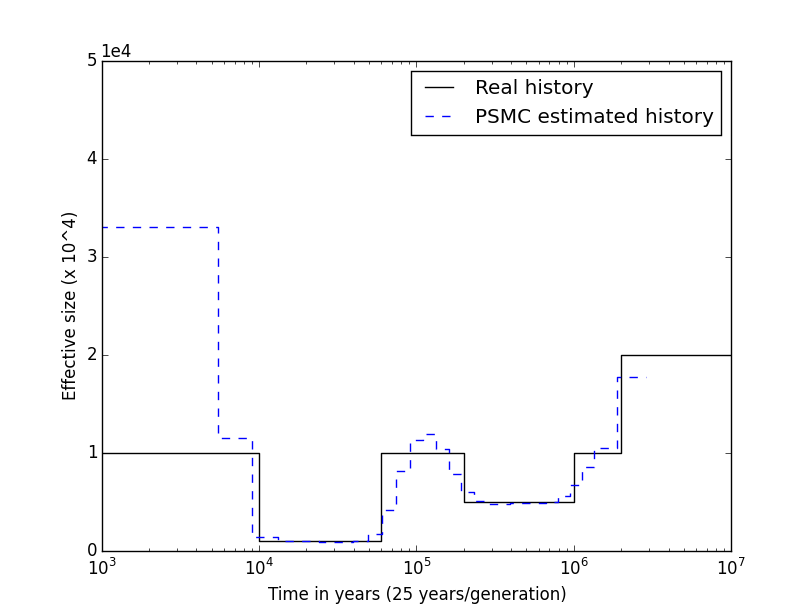
You can also plot the psmc results by using the perl script utils/psmc_plot.pl wich comes with the PSMC software.
[1] http://www.nature.com/articles/nature10231 "Heng Li & Richard Durbin, Inference of human population history from individual whole-genome sequences, Nature 475, 493–496 (28 July 2011) doi:10.1038/nature102312011"
Back
Back to Day #4.
Back to first page.