Parsing data records II
In many cases you will need to compare data from different files. As an example, consider the two files:
1) A Uniprot multiple sequence FASTA file SwissProt-Human.fasta, available here.
SwissProt-Human.fasta
>sp|P31946|1433B_HUMAN 14-3-3 protein beta/alpha OS=Homo sapiens
MTMDKSELVQKAKLAEQAERYDDMAAAMKAVTEQGHELSNEERNLLSVAYKNVVGARRSS
WRVISSIEQKTERNEKKQQMGKEYREKIEAELQDICNDVLELLDKYLIPNATQPESKVFY
LKMKGDYFRYLSEVASGDNKQTTVSNSQQAYQEAFEISKKEMQPTHPIRLGLALNFSVFY
YEILNSPEKACSLAKTAFDEAIAELDTLNEESYKDSTLIMQLLRDNLTLWTSENQGDEGD
AGEGEN
>sp|P62258|1433E_HUMAN 14-3-3 protein epsilon OS=Homo sapiens
MDDREDLVYQAKLAEQAERYDEMVESMKKVAGMDVELTVEERNLLSVAYKNVIGARRASW
RIISSIEQKEENKGGEDKLKMIREYRQMVETELKLICCDILDVLDKHLIPAANTGESKVF
YYKMKGDYHRYLAEFATGNDRKEAAENSLVAYKAASDIAMTELPPTHPIRLGLALNFSVF
YYEILNSPDRACRLAKAAFDDAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSDMQGDGE
EQNKEALQDVEDENQ
>sp|Q04917|1433F_HUMAN 14-3-3 protein eta OS=Homo sapiens GN=YWHAH
MGDREQLLQRARLAEQAERYDDMASAMKAVTELNEPLSNEDRNLLSVAYKNVVGARRSSW
RVISSIEQKTMADGNEKKLEKVKAYREKIEKELETVCNDVLSLLDKFLIKNCNDFQYESK
VFYLKMKGDYYRYLAEVASGEKKNSVVEASEAAYKEAFEISKEQMQPTHPIRLGLALNFS
VFYYEIQNAPEQACLLAKQAFDDAIAELDTLNEDSYKDSTLIMQLLRDNLTLWTSDQQDE
EAGEGN
...
...
...
2) A file cancer-expressed.txt containing a list of protein accession numbers (ACs), available here:
Q5XXA6
Q9Y5P2
Q14667
O75387
Q8WV07
Q8CH62
Q9GZY1
Q9NQQ7
Q8VCX2
Q7Z769
In order to compare the content of the two files, you might first want to store the 10 Uniprot ID in a data structure. In practice:
- Read 10 SwissProt ACs from the file
- Store them in a data structure
Lists are nice and veeeeery flexible data structures
['Q5XXA6', 'Q9Y5P2', 'Q14667', 'O75387', 'Q8WV07','Q8CH62', 'Q9GZY1', 'Q9NQQ7', 'Q8VCX2', 'Q7Z769']
List data structure
A list is a mutable ordered collection of objects
The elements of a list can be any kind of object:
- numbers
- strings
- tuples
- lists
- dictionaries
- function calls
- etcetera
L = [1, [2,3], 4.52, 'DNA']
L = [] # the empty list
Operations with lists
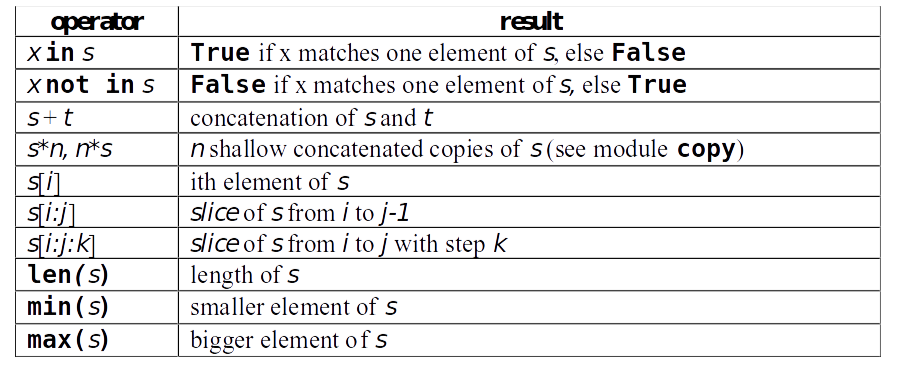
>>> L = [1,"hello",12.1,[1,2,"three"],"seq",(1,2)]
>>> L[0] # indexing
1
>>> L[3] # indexing
[1, 2, 'three']
>>> L[3][2] # indexing
'three'
>>> L[-1] # negative indexing
(1, 2)
>>> L[2:4]
[12.1, [1, 2, 'three']] # slicing
>>> L[2:]
[12.1, [1, 2, 'three'], 'seq', (1, 2)] #slicing shorthand
>>>
The elements of a list can be changed/replaced after the list has been defined These operations CHANGE the list
| l[i] = x |
| l[i:j] = t |
| del l[i:j] |
| del l[i:j:k] |
| l.append(x) |
| l.extend(s) |
s = any sequence
>>> l = [2,3,5,7,8,['a','b'],'a','b','cde']
>>> l[0] = 1
>>> l
[1, 3, 5, 7, 8, ['a', 'b'], 'a', 'b', 'cde']
>>> l[0:3] = 'DNA'
>>> l
['D', 'N', 'A', 7, 8, ['a', 'b'], 'a', 'b', 'cde']
>>> del l[0:5]
>>> l
[['a', 'b'], 'a', 'b', 'cde']
>>> l.append('DNA')
>>> l
[['a', 'b'], 'a', 'b', 'cde', 'DNA']
>>> l.extend('dna')
>>> l
[['a', 'b'], 'a', 'b', 'cde', 'DNA', 'd', 'n', 'a']
>>>
The elements of a list can be changed/replaced after the list has been defined
| l.count(x) |
| l.index(x) |
| l.insert(i, x) |
| l.pop(i) |
| l.remove(x) |
>>> l = [1,3,5,7,8,['a','b'],'a','b','cde']
>>> l.count('a')
>>> l
1
>>> l.index(8)
4
>>> l.insert(4, 80)
>>> l
[1, 3, 5, 7, 80, 8, ['a', 'b'], 'a', 'b', 'cde']
>>> l.pop(4)
80
>>> l
[1, 3, 5, 7, 8, ['a', 'b'], 'a', 'b', 'cde']
>>> l.pop()
'cde'
>>> l
[1, 3, 5, 7, 8, ['a', 'b'], 'a', 'b']
>>> l.remove(8)
[1, 3, 5, 7, ['a', 'b'], 'a', 'b']
The elements of a list can be changed/replaced after the list has been defined
| l.reverse() |
| l.sort() |
| sorted(l) |
>>> l = [4, 3, 2, 1, 5, 6, 7, 8]
>>> l.reverse()
>>> l
[8, 7, 6, 5, 1, 2, 3, 4]
>>> new = sorted(l)
>>> new
[1, 2, 3, 4, 5, 6, 7, 8]
>>> l
[8, 7, 6, 5, 1, 2, 3, 4]
>>> l.sort()
>>> l
[1, 2, 3, 4, 5, 6, 7, 8]
Putting together lists and loops
range() and xrange() built-in functions
>>> range(10)
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9]
>>> range(1, 11)
[1, 2, 3, 4, 5, 6, 7, 8, 9, 10]
>>> range(0, 30, 5)
[0, 5, 10, 15, 20, 25]
>>> range(0, 10, 3)
[0, 3, 6, 9]
>>> range(0, -10, -1)
[0, -1, -2, -3, -4, -5, -6, -7, -8, -9]
>>> range(0)
[]
>>> range(1, 0)
[]
# the xrange()method is more commonly used in for loops than range()
>>>for i in xrange(5):
… print i
…
0,1,2,3,4
The xrange() method generates the values upon call, i.e. it does not
store them into a variable
Indexing
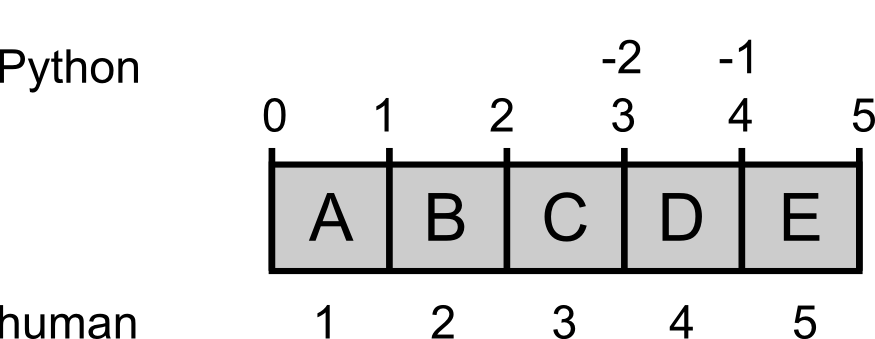
Computers treat an address in memory as the starting point of a body of data. In the same sense, an index in Python always refers to such a starting point, something that is in between two objects in memory. We humans in contrast always count the objects themselves.
Challenge #1
Download the file
cancer-expressed.txtform here
- Read 10 SwissProt ACs from a file
- Store them into a list
- Print the list
See the Solution to challenge #1
Challenge #2
Download the Uniprot multiple sequence FASTA file
SwissProt-Human.fastahere.
- Create a list containing Uniprot ACs extracted from the FASTA file
- Print the list
See the Solution to challenge #2
Challenge #3
Download the Uniprot multiple sequence FASTA file
SwissProt-Human.fastafrom here. Download the filecancer-expressed.txtcontaining a list of ACs from here
- Read the ACs and store them in a pyton structure
- Read the human FASTA file one record after the other. Check if the record header contains one of the 10 ACs.
- If YES, copy the header to a new file.
See the Solution to challenge #3
Challenge #4
Download the Uniprot multiple sequence FASTA file
SwissProt-Human.fastafrom here. Download the filecancer-expressed.txtcontaining a list of ACs from here.
- Read the ACs and store them in a pyton structure
- Read the human FASTA file one record after the other. Check if the record header contains one of the 10 ACs.
- If YES, copy the header and the sequence to a new file.
See the Solution to challenge #4
Putting together conditions and loops while loops
The while statement is used for executing a set of statements until a given condition is met
**while <condition 1>:
<statements 1>**
>>> a = 1
>>> while a < 5:
... print a,
... a = a + 1
BUT:
>>> a = 1
>>> while a > 0:
... if a == 5: break
... print a,
... a = a + 1
... else: print "loop terminated"
...
1 2 3 4
The Boolean values True and False
if and while statements return a False value when they are applied to:
- None
- 0
- Empty data structures: ‘’,(),[],{}
The statements in an if or a while block are executed only if the condition returns the value True.
>>> p = 'protein'
>>> if p: print 'True'
...
True
>>> n = 0
>>> while 1:
... print n,
... n = n + 1
... if n > 5: break
...
0 1 2 3 4 5
>>>
We can use while loops to read files (but usually we won’t do it):
cancer_file = open('cancer-expressed.txt')
cancer_list = []
line = cancer_file.readline()
while line:
AC = line.strip()
cancer_list.append(AC)
line = cancer_file.readline()
Back
Back to main page.