Repeating things
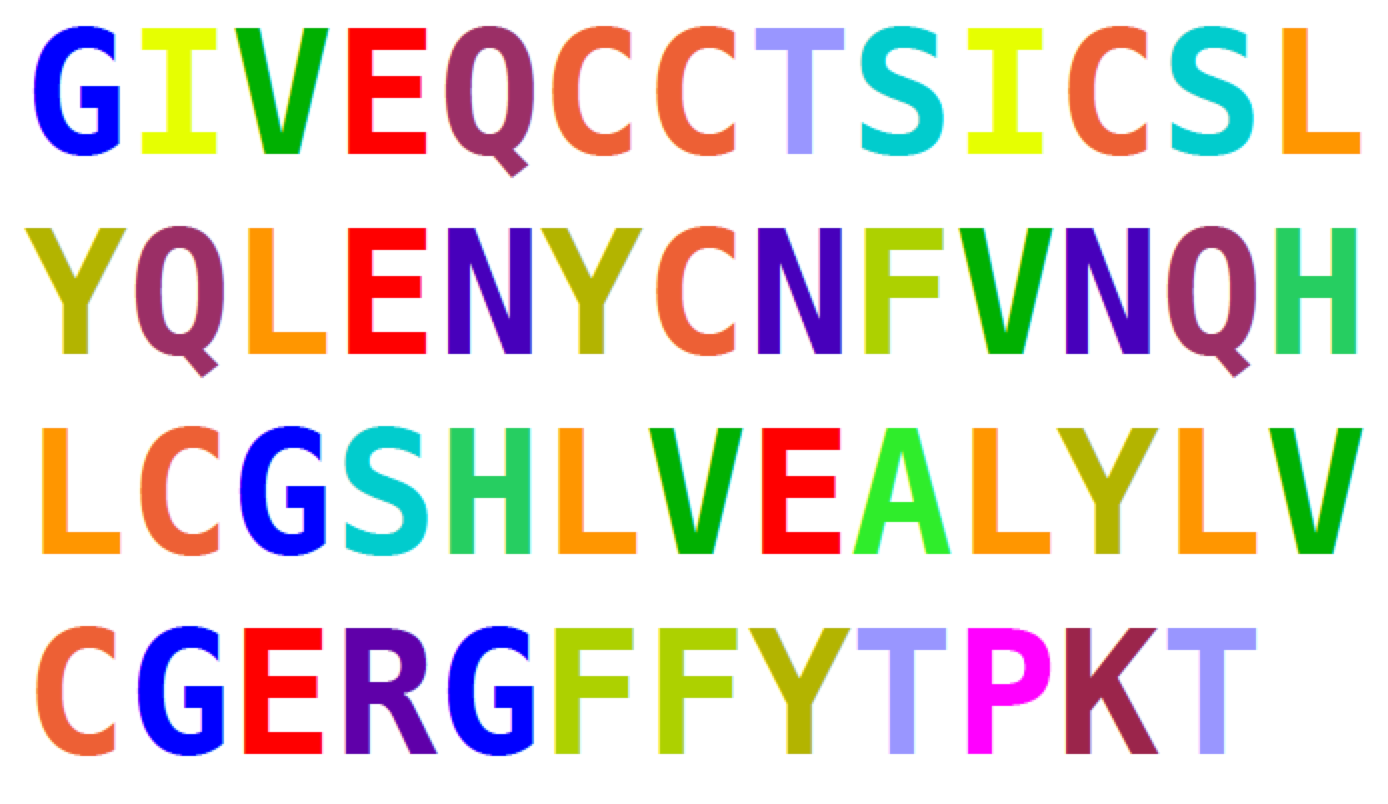
Counting amino acids
# insulin [Homo sapiens] GI:386828
insulin = "GIVEQCCTSICSLYQLENYCFVNQHLC\
GSHLVEALYLVGERGFFYTPKT"
for amino_acid in "ACDEFGHIKLMNPQRSTVWY":
number = insulin.count(amino_acid)
print amino_acid, number
Repetitive tasks
Consider the solutions of challenge #4 from previous session:
dna = "AGCTTCGA"
print dna.count("A")
print dna.count("C")
print dna.count("T")
print dna.count("G")
dna = "AGCTTCGA"
elem = dna.count ("A")
print elem
elem = dna.count ("C")
print elem
elem = dna.count ("T")
print elem
elem = dna.count ("G")
print elem
##Loops with for
The for command repeats other commands:
dna = "AGCTTCGA”
for base in "ACTG":
print dna.count(base)
The commands that are repeated must be indented (shifted right by four spaces).
Compare
dna = "AGCTTCGA”
for base in "ACTG":
print dna.count(base)
Would you prefer this implementation?
dna = "AGCTTCGA"
print dna.count("A")
print dna.count("C")
print dna.count("T")
print dna.count("G")
Why or why not?
Challenge #1
Retrieve the 1132-residue sequence of human telomerase reverse transcriptase isoform 1 from the NCBI protein database. Choose the FASTA format. Copy the sequence to a text file (
telomerase.txt). Write a program that reads the telomerase.txt file and prints first the whole sequence and then the sequence residue by residue.
See the Solution to challenge #1
Use a for loop to read a file line by line
Input_file = open(“my_file.txt”)
for line in Input_file:
print line
Look how beautiful it can be…
import urllib
url = 'http://www.uniprot.org/\
uniprot/P12931.fasta'
src_human = urllib.urlopen(url)
for line in src_human:
print line,
Challenge #2
Write a file and program that reads the
telomerase.txtprints its content line by line.
See the Solution to challenge #2
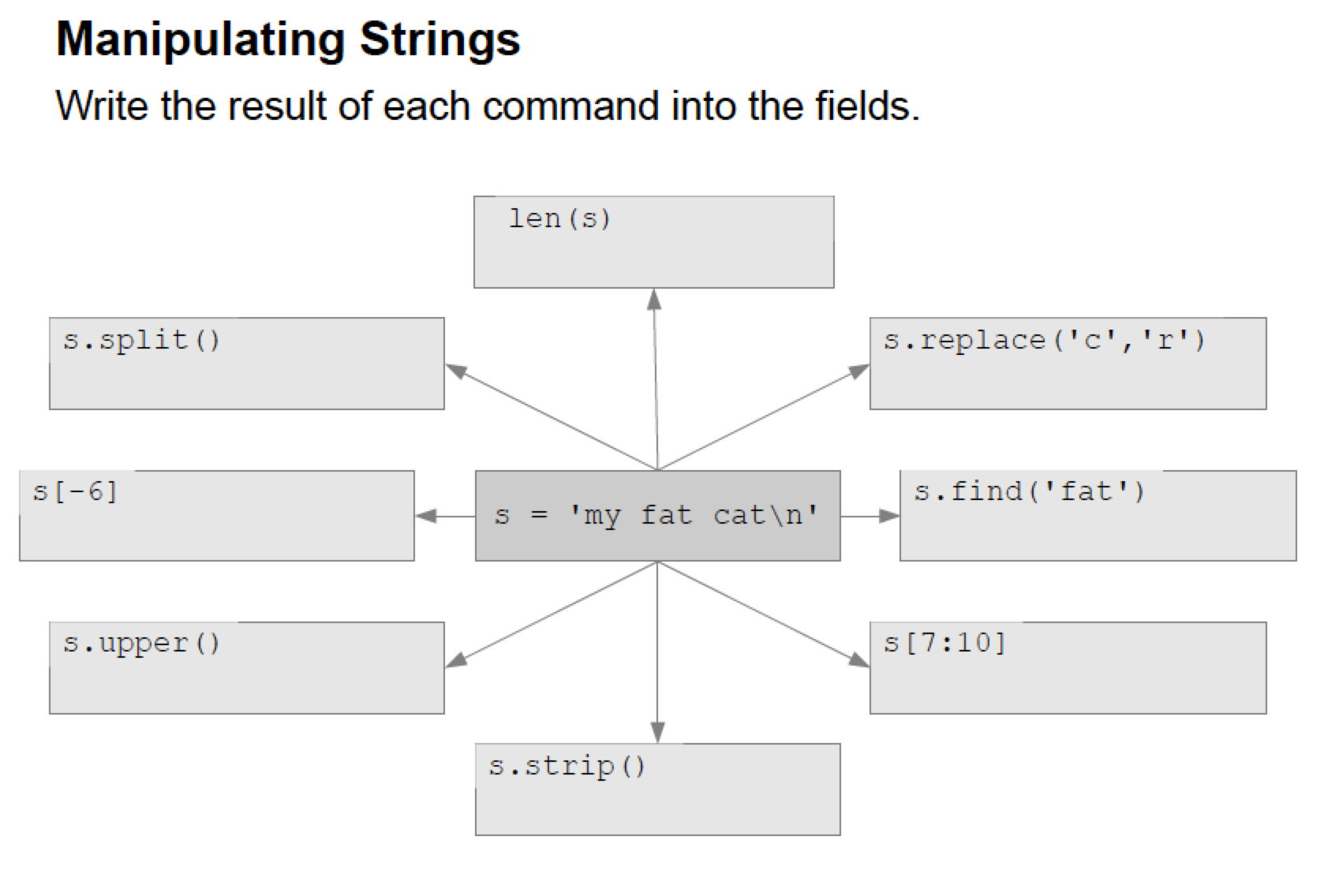
Manipulating string
Challenge #3
Which amino acid is the most frequent in the sequence of the telomerase reverse transcriptase isoform 1?
See the Solution to challenge #3
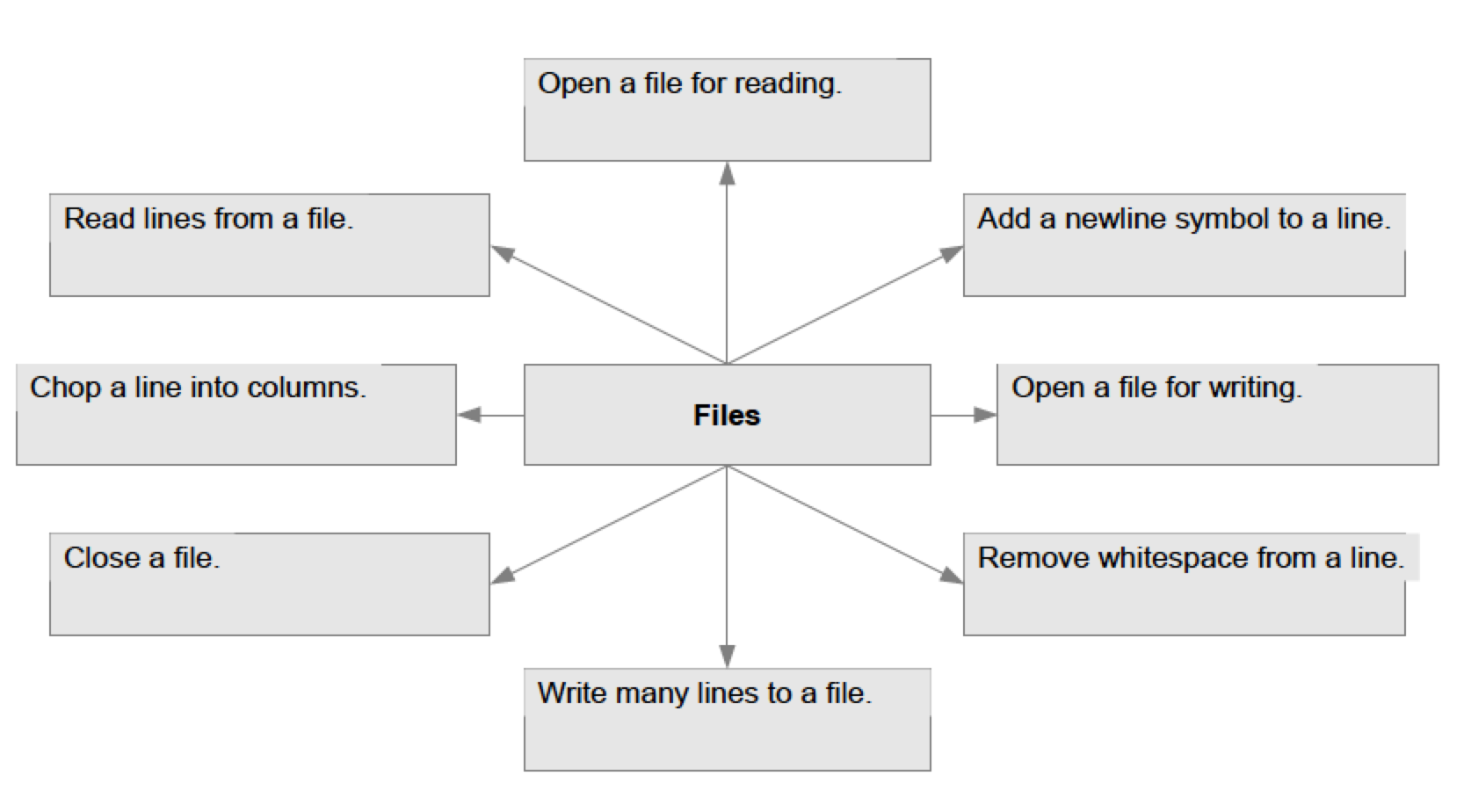
Recap
Back
Back to first page.