Proteogeonomics
Proteogenomics is the scientific field at the interface between proteomics and genomics (1). First, proteogenomic applications focused on improving genomic annotation through the use of proteomics data (2). Over the years, proteogenomics rapidly developed as a rich scientific field for the joint analysis of genomic and proteomic information, generating fundamental knowledge on the interplay between genome and proteome.
Due to the novelty of the field, the complexity of the analyses, and the volume of data involved, proteogenomics poses multiple bioinformatic challenges (3). These tutorials present methods that can be used to analyse results from proteogenomic pipelines.
Introduction slides are available here
A word of caution
Please note that this tutorial starts from the results of proteomic and genomic analyses, and does not cover the details of respective bioinformatic pipelines, like the calling of variants. Please note also that the generation and analysis of proteomic data for proteogenomic application differ from standard proteomic analyses, requiring specific methods and expertise. Notably, the identification of non-canonical genetic products of transcription/translation and the calling of variants in proteomics data require high coverage and high resolution proteomic analyses. These analyses also require the use of complex bioinformatic workflows in high performance environments. Finally, they can benefit from tailoring the experimental workflow in relationship to the bioinformatic analysis (4).
Note that establishing a proteogenomic workflow, in both wet- and dry-lab is a long-term endeavor. If you plan to conduct a proteogenomic experiment, please give particular attention to the following questions:
- Do I have enough sample material, replicates, and controls?
- Has the proteomics lab sufficient instrumentation and expertise to run the samples?
- Is the proteogenomic bioinformatic pipeline in place and able to handle the volume of data generated by my experiment?
When in doubt, consider running a small pilot experiment to benchmark the performance of your analytical set-up. Also, prioritize proteomics labs that have experience with proteogenomic analyses and analyzing the data.
The present tutorials focus on human samples and resources. While the methods presented are generic, differences are to be expected when working with other organisms, notably on the level of genomic and proteomic data annotation. Please note that working with multiple organisms, e.g. in metaproteogenomic analyses (5), is yet another level of complexity, posing multiple challenges for the interpretation of the data (6).
Proteogenomics Lexicon
Analysing proteogenomics data requires familiarity with both genomic and proteomic fields, which can leave scientists discombobulated. We provide here a short lexicon of the terms and abbreviations the reader will encounter in the literature. Please do not hesitate to suggest terms that should be added in our issue tracker.
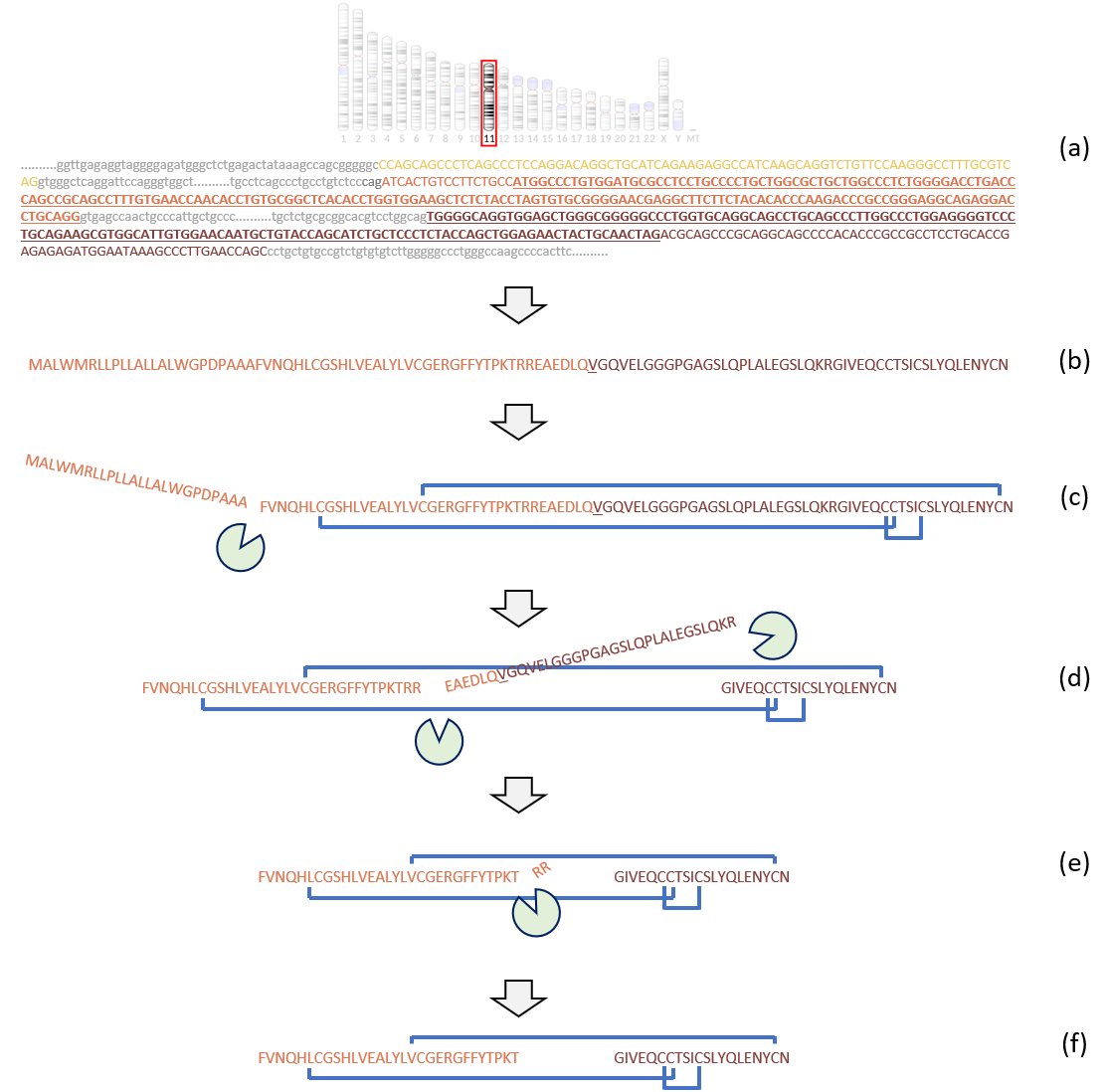
The production of human insulin from the transcript INS-201 according to Ensembl release 97. (a) The transcript is coded by three exons on chromosome eleven, colored in yellow, orange, and dark red. The translated sequence is underlined. Sequence variation in the translated sequence can result in sequence variations in the amino acid sequence, and hence different proteoforms. (b) The sequence obtained after translation represents the raw proteoform of insulin, called preproinsulin, which requires posttranslational maturation to obtain the mature form of insulin. Amino acids are colored according to the coding exons and the residue overlapping splice site is underlined. (c) The signal peptide is cleaved, yielding proinsulin, and cysteines cross-linked by disulphide bonds, making a new proteoform of insulin. (d) Proteases cleave a large fragment of the sequence, the C-peptide. The C-peptide is often used as a proxy to measure insulin production. (e) Proteases cleave pairs of amino-acids, yielding the mature form of insulin. (f) The mature form of insulin consists of two cross-linked peptides. It can be further modified, yielding even more proteoforms. From (7), adapted from en.wikipedia.org/wiki/Insulin.
-
Variant, Mutation, Alteration: A genetic variant or mutation refers to a variation in the genetic sequence. When the variation is not inherited, it is called alteration. Variants involving the substitution of a single nucleotide are called single-nucleotide polymorphism (SNP) or single-nucleotide variant (SNV). SNP refers to variants where each version is carried by more than 1% of the population. When a variation of one amino acid is detected in the proteome, it is referred to as single amino acid variant (SAV or SAAV). Variants involving the deletion or insertion of genetic code are called indels. Variants involving the repeat of sections of the genome are examples of structural variation, and called Copy-number variation (CNV). When the variation is not inherited, it is called copy-number alteration (CNA).
-
Peptides, Proteins, isoforms, and proteoforms: peptides and proteins are short and long chains of amino acids, respectively. Proteins are generally associated to a gene which codes their amino acid chain. Differences in splicing yield different isoforms for most proteins. During their lifetime, proteins undergo structural modification: cleavage, folding, cross-linking, post-translational modification (PTM), etc. These modifications yield very different forms for each protein, called proteoforms (8).
-
Non-canonical genetic product: Peptide or protein produced from regions of the genome that are canonically non-coding.
Legend
You will find the following icons throughout the text:
- :pencil2: : You need to do something
- :speech_balloon: : Something you might want to discuss with someone else (or yourself).
- :thought_balloon: : A question to trigger your attention on an important point of detail. Clicking the icon takes you to the answer and more literature on the subject. Clicking the icon in the answer takes you back to the question.
Please note that questions rarely trigger a yes/no answer. The answers provided represent important elements to take into account when analyzing data, but are by no means exhaustive or universal.
Processing
This tutorial is organized in notebooks that contains R code that can be run directly from the Rmd file. It assumes that the R working directory is the proteogenomics folder of the repository, e.g. /myfolder/IBIP19/pages/proteogenomics. We recommend using RStudio to run this tutorial.
Libraries
You will need the following libraries, please make sure that they are installed.
- We will use tidyr to import data, we recommend this cheat sheet.
- We will use dplyr to transform data, we recommend this cheat sheet.
- We will use ggplot2 to plot data, we recommend this cheat sheet.
- We will use gtable to organize plots.
- We will use gamlss for normalization.
- We will use mclust for Gaussian mixture modeling.
- We will use igraph for graph manipulation.
- We will use conflicted to manage conflict resolution to function names between packages.
Warning: conflicted is not available in cran yet, you will need to install it using devtools. See the installation instructions for more details.
Tutorials
-
1. Novel peptides: mapping proteomics results to non-coding regions of the genome.
-
2. Variation Analysis: studying sequence and splicing variation in proteomics data.
-
3. CNA-Protein: linking structural variants and protein levels to study CNA dampening/silencing.
-
4. RNA-Protein: comparing RNA and protein levels to identify key biological mechanisms.
References
(1) Proteogenomics: concepts, applications and computational strategies
(2) Proteogenomic mapping as a complementary method to perform genome annotation
(3) Proteogenomics from a bioinformatics angle: A growing field
(4) HiRIEF LC-MS enables deep proteome coverage and unbiased proteogenomics
(7) Proteomics in Processing Metabolomics and Proteomics Data