- Introduction
- 1. Background
- 2. Retrieving sequences
- 3. Sequence alignment
- 4. Distance-based analyses
- 5. Recombination
- 6. Maximum likelihood based analyses
- 7. Visualising trees
- 8. Time-stamped phylogenies
- 9. Bayesian reconstruction of time trees
- 10. Effective population size estimation
- 11. Structured populations
- 12. References
- Published with GitBook
Distances walkthrough
First load the libraries you need.
library(ape)
library(phangorn)
library(network)
## network: Classes for Relational Data
## Version 1.12.0 created on 2015-03-04.
## copyright (c) 2005, Carter T. Butts, University of California-Irvine
## Mark S. Handcock, University of California -- Los Angeles
## David R. Hunter, Penn State University
## Martina Morris, University of Washington
## Skye Bender-deMoll, University of Washington
## For citation information, type citation("network").
## Type help("network-package") to get started.
Load the multiple sequence alignment using read.dna from the ape library.
myalignment.filename <- "ray2000_pruned_degapped.fas"
myalignment <- read.dna(myalignment.filename,format="fasta",as.matrix=TRUE)
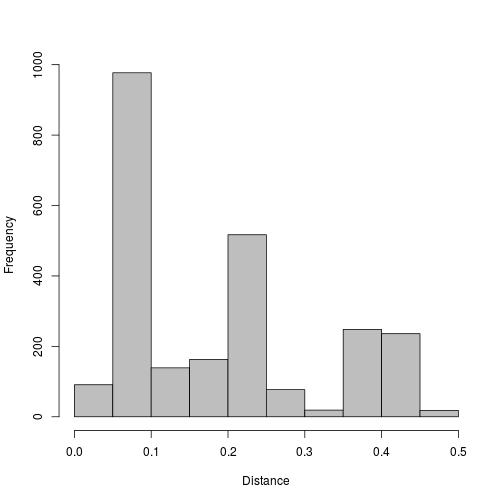
Calculate distances using the TN93 model.
myalignment.dist.tn93 <- dist.dna(myalignment,model="TN93",as.matrix=TRUE)
Plot out the distances.
hist(myalignment.dist.tn93[lower.tri(myalignment.dist.tn93)],xlab="Distance",ylab="Frequency",main="",col="grey")
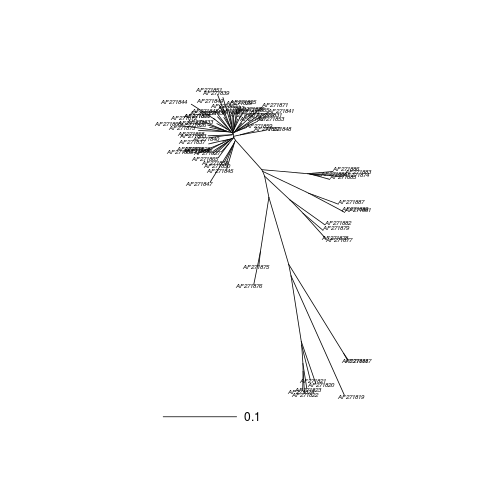
Calculate a neighbour joining tree.
myalignment.tn93.nj <- nj(myalignment.dist.tn93)
Plot the tree out and add a scale bar.
plot(myalignment.tn93.nj,"unrooted",cex=0.5)
add.scale.bar(length=0.1)
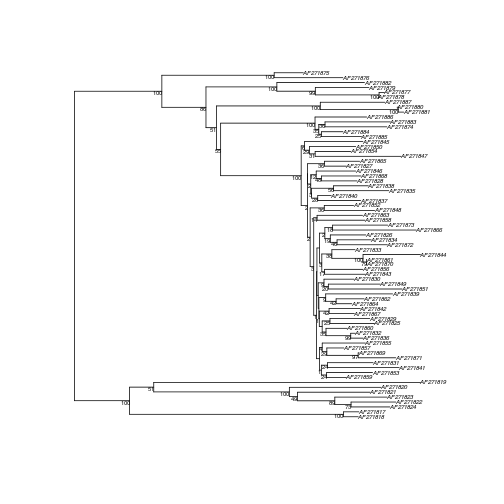
Bootstrap the alignment; we first define a function to make a tree from the alignment, then call boot.phylo.
maketree <- function(x) nj(dist.dna(x,model="TN93"))
myalignment.tn93.nj.boot <- boot.phylo(myalignment.tn93.nj,myalignment,maketree,B=100,block=3,trees=TRUE,quiet=FALSE,rooted=FALSE)
##
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|=== | 5%
|
|==== | 6%
|
|===== | 7%
|
|===== | 8%
|
|====== | 9%
|
|====== | 10%
|
|======= | 11%
|
|======== | 12%
|
|======== | 13%
|
|========= | 14%
|
|========== | 15%
|
|========== | 16%
|
|=========== | 17%
|
|============ | 18%
|
|============ | 19%
|
|============= | 20%
|
|============== | 21%
|
|============== | 22%
|
|=============== | 23%
|
|================ | 24%
|
|================ | 25%
|
|================= | 26%
|
|================== | 27%
|
|================== | 28%
|
|=================== | 29%
|
|==================== | 30%
|
|==================== | 31%
|
|===================== | 32%
|
|===================== | 33%
|
|====================== | 34%
|
|======================= | 35%
|
|======================= | 36%
|
|======================== | 37%
|
|========================= | 38%
|
|========================= | 39%
|
|========================== | 40%
|
|=========================== | 41%
|
|=========================== | 42%
|
|============================ | 43%
|
|============================= | 44%
|
|============================= | 45%
|
|============================== | 46%
|
|=============================== | 47%
|
|=============================== | 48%
|
|================================ | 49%
|
|================================ | 50%
|
|================================= | 51%
|
|================================== | 52%
|
|================================== | 53%
|
|=================================== | 54%
|
|==================================== | 55%
|
|==================================== | 56%
|
|===================================== | 57%
|
|====================================== | 58%
|
|====================================== | 59%
|
|======================================= | 60%
|
|======================================== | 61%
|
|======================================== | 62%
|
|========================================= | 63%
|
|========================================== | 64%
|
|========================================== | 65%
|
|=========================================== | 66%
|
|============================================ | 67%
|
|============================================ | 68%
|
|============================================= | 69%
|
|============================================== | 70%
|
|============================================== | 71%
|
|=============================================== | 72%
|
|=============================================== | 73%
|
|================================================ | 74%
|
|================================================= | 75%
|
|================================================= | 76%
|
|================================================== | 77%
|
|=================================================== | 78%
|
|=================================================== | 79%
|
|==================================================== | 80%
|
|===================================================== | 81%
|
|===================================================== | 82%
|
|====================================================== | 83%
|
|======================================================= | 84%
|
|======================================================= | 85%
|
|======================================================== | 86%
|
|========================================================= | 87%
|
|========================================================= | 88%
|
|========================================================== | 89%
|
|========================================================== | 90%
|
|=========================================================== | 91%
|
|============================================================ | 92%
|
|============================================================ | 93%
|
|============================================================= | 94%
|
|============================================================== | 95%
|
|============================================================== | 96%
|
|=============================================================== | 97%
|
|================================================================ | 98%
|
|================================================================ | 99%
|
|=================================================================| 100%
## Calculating bootstrap values... done.
Now we can add the bootstrap supports to the tree.
plotBS(myalignment.tn93.nj,myalignment.tn93.nj.boot$trees,type="phylogram",cex=0.5)
We can also create a network of 'similar' sequences, by defining a threshold distance.
thresh <- 0.01
myalignment.dist.tn93.net <- network(myalignment.dist.tn93<=thresh)
plot(myalignment.dist.tn93.net)