- Introduction
- 1. Background
- 2. Retrieving sequences
- 3. Sequence alignment
- 4. Distance-based analyses
- 5. Recombination
- 6. Maximum likelihood based analyses
- 7. Visualising trees
- 8. Time-stamped phylogenies
- 9. Bayesian reconstruction of time trees
- 10. Effective population size estimation
- 11. Structured populations
- 12. References
- Published with GitBook
Background
- In the context of viruses, molecular epidemiology is used to describe how we can make inferences about the transmission, distribution, etiology, and prevention of viral infections within a population
- Given the decreasing cost of sequencing, the use of molecular epidemiology to understand viruses is increasing
Questions
- There are many questions that can be addressed with sequence data
- When did an epidemic start?
- How did an epidemic spread spatially, and between different risk groups?
- What are the dynamics of transmission over time
- These questions involve reconstructing phylogenetic trees from sequence data
Transmission and phylogenies
Ideally, we would like to know about the transmission history of a pathogen, but even in ideal cases, there is a loss of information in the phylogeny.
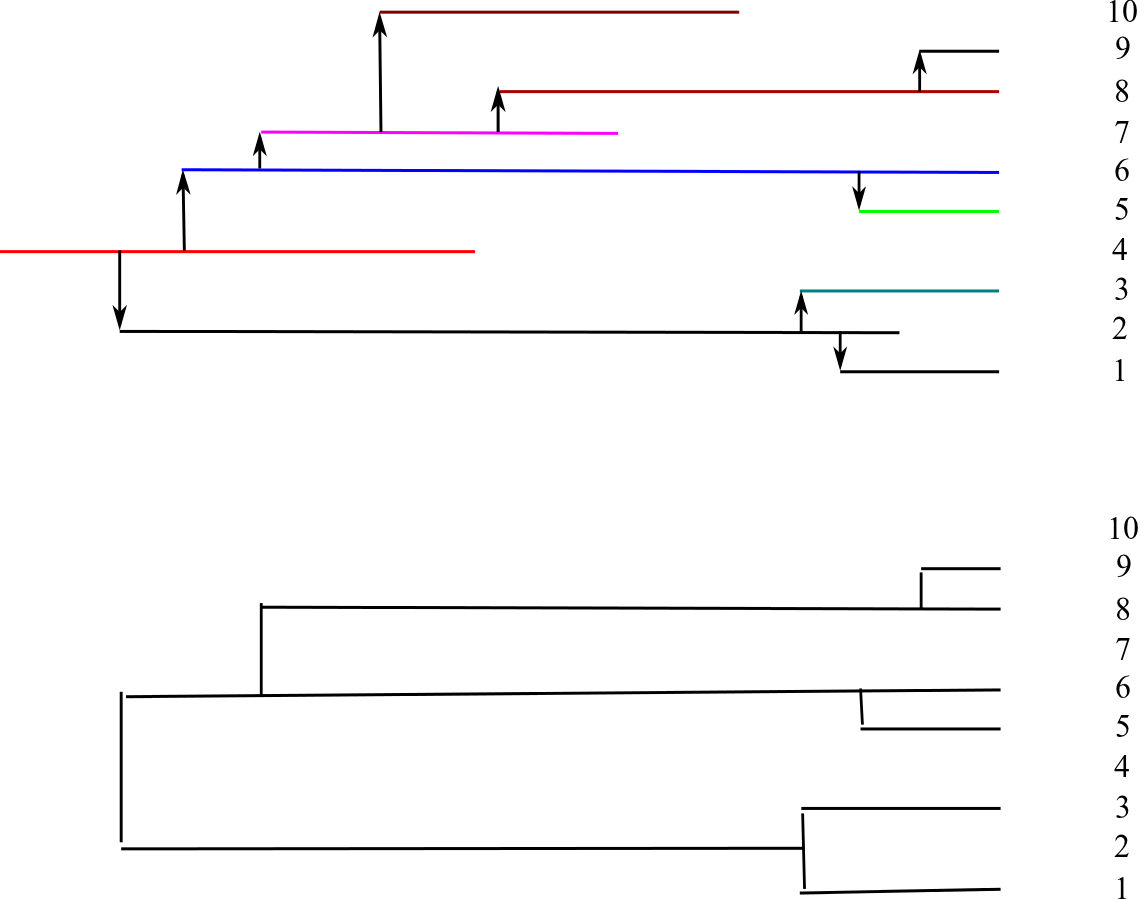
- No information before the common ancestor of the samples
- Direction of transmission is lost
- No information on individuals who have died/recovered before sampling
- We do not know which host is infected by which virus (except sampled individuals)
- Not all infection events are 'observed'
Workflow
- A typical molecular epidemiology workflow is rather linear
Problems
- There are many steps
- There are many software packages available to perform even a single part of this workflow
Why R?
- Free, general purpose statistical software
- Many libraries (>4000), including those for sequence analysis
- Can call external programs
- Literate programming
Why RStudio
- R runs in a terminal
- RStudio sits on top of R, and offers a number of additional features
- Multiple windows for editing and running of code
- Workspace browser
- File browser
- Integrated help
- Graphics window
What you'll (hopefully) learn
- Retrieving sequence data
- Obtaining sequence metadata
- Processing and altering sequence names
- Developing simple pipelines
What you won't learn
- Processing of next-generation sequencing data
- BEAST
Course structure
- Overall structure borrowed from The Phylogenetic Handbook, Second Edition
- Brief theoretical background
- Introduction to the practical implementation
- Worked example
- Exercises
Next steps
Next, we'll go through a minimal example of generating a phylogeny. We'll do much more, both in terms of upstream analysis (data processing) and in downstream analysis (visualisation and interpretation) later.